Single-cell epigenetic regulatory landscape in mammalian perinatal testis development
Cell Diversity in the Testis
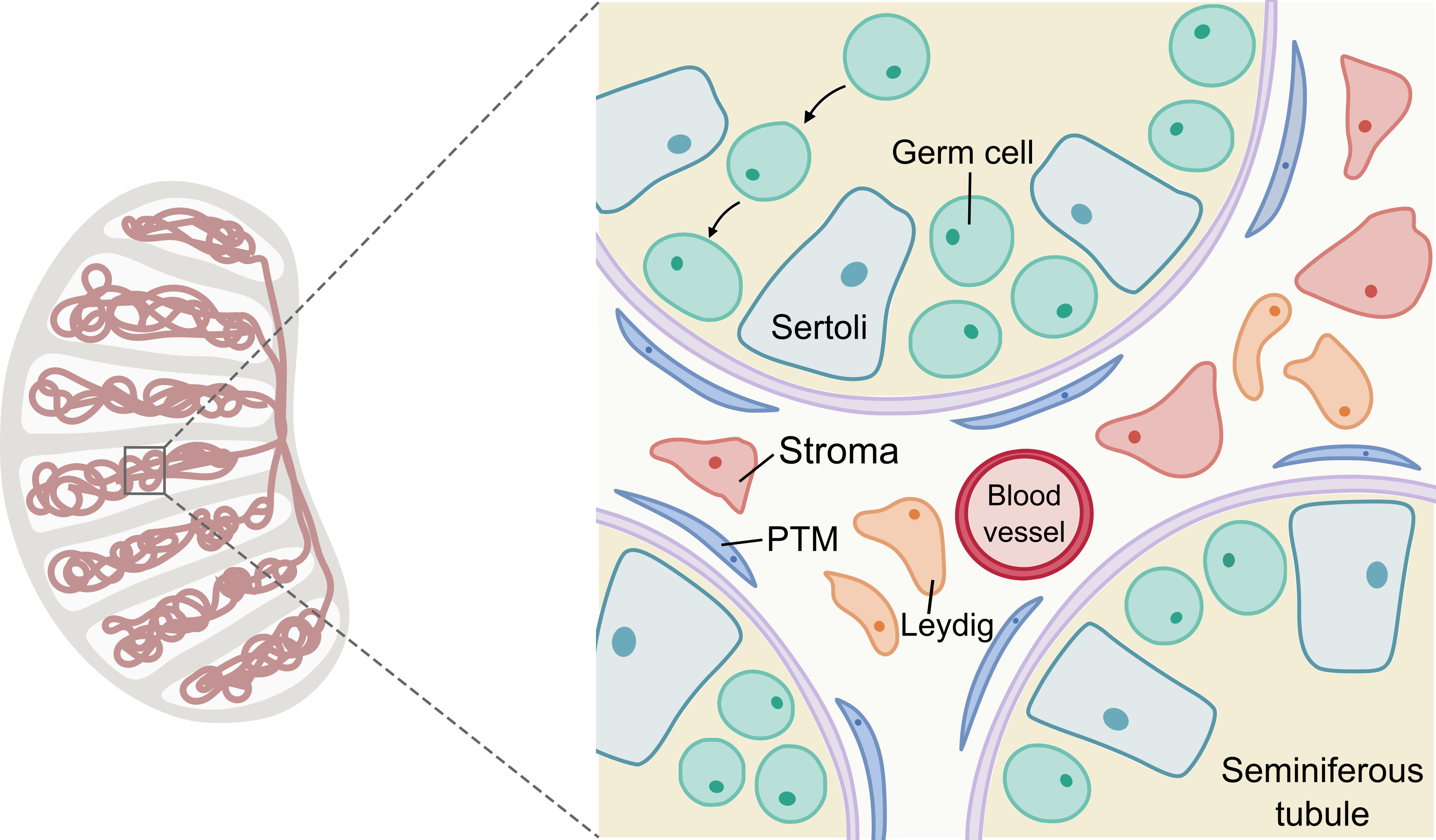
Mammalian testis consists of germ cells and different somatic cell types to maintain spermatogenesis and fertility. While germ cells give rise to sperms, specialized somatic cells are crucial for maintaining normal germ cell development and spermatogenesis. Sertoli cells nourish the germ cells inside the seminiferous tubules. Outside the seminiferous tubules, the interstitial compartment contains stroma and various cell types with distinct functions. PTMs are thin smooth muscle cells that can facilitate the movement of sperm and secrete extracellular matrix materials. Leydig cells are responsible for steroidogenesis and spermatogenesis. Testicular tissue-resident macrophages are involved in steroidogenesis, spermatogonia differentiation and Leydig cell function.
What is single-cell ATAC-Seq?
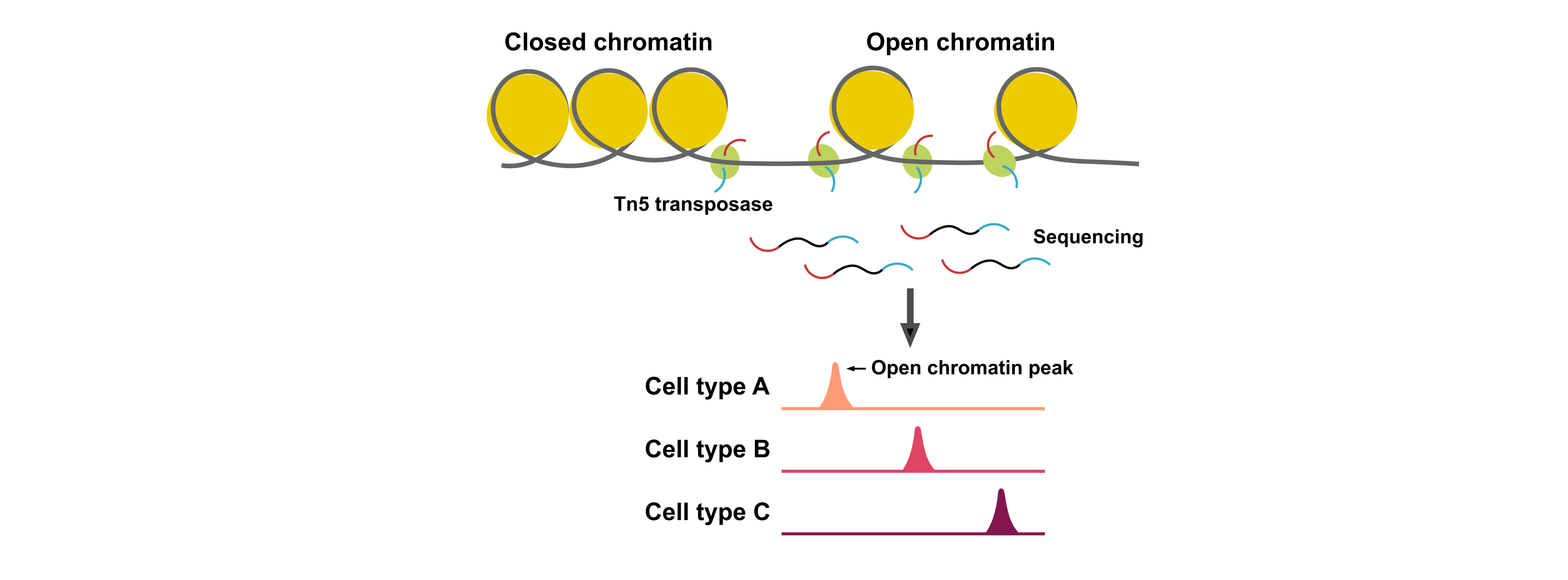
ATAC-Seq (Assay for Transposase-Accessible Chromatin) is an assay for profiling genome-wide chromatin accessibility. To initiate gene expression, transcription factors bind to a specific DNA sequence. After the binding, the chromatin will become more open and accessible. By ATAC-seq, If a part of the genome is accessible, the Tn5 transposase will make a cut on the accessible chromatin. Through sequencing the open chromatin region, we can gain information on TF dynamics and cis-regulatory elements. By profiling the chromatin landscapes at single-cell level, single cell ATAC-Seq (scATAC-Seq) can deconvolve complex cell populations, discover regulatory elements and map regulatory dynamics during complex cellular processes.
Deconvolving cell populations in testis
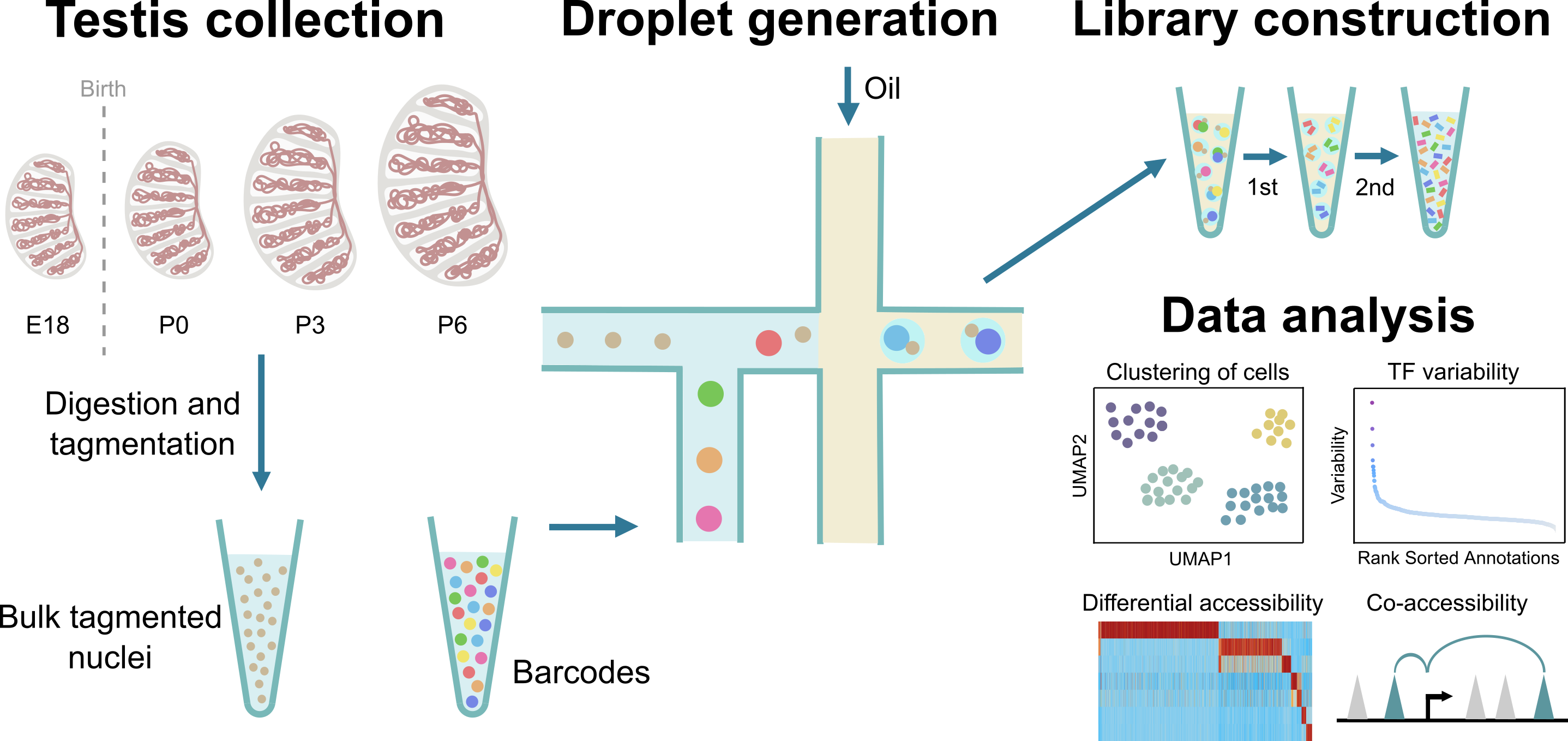
Testicular cells display extensive developmental dynamics during the perinatal period. To delineate these dynamic changes, we profiled the chromatin accessibility landscapes of mouse perinatal testis across embryonic and postnatal stages by scATAC-Seq. Altogether, we profiled chromatin accessibility in 25,613 individual cells. The dataset identified key cell type-specific TFs, defined the cellular differentiation trajectory, and characterized cell type-specific chromatin interaction networks.
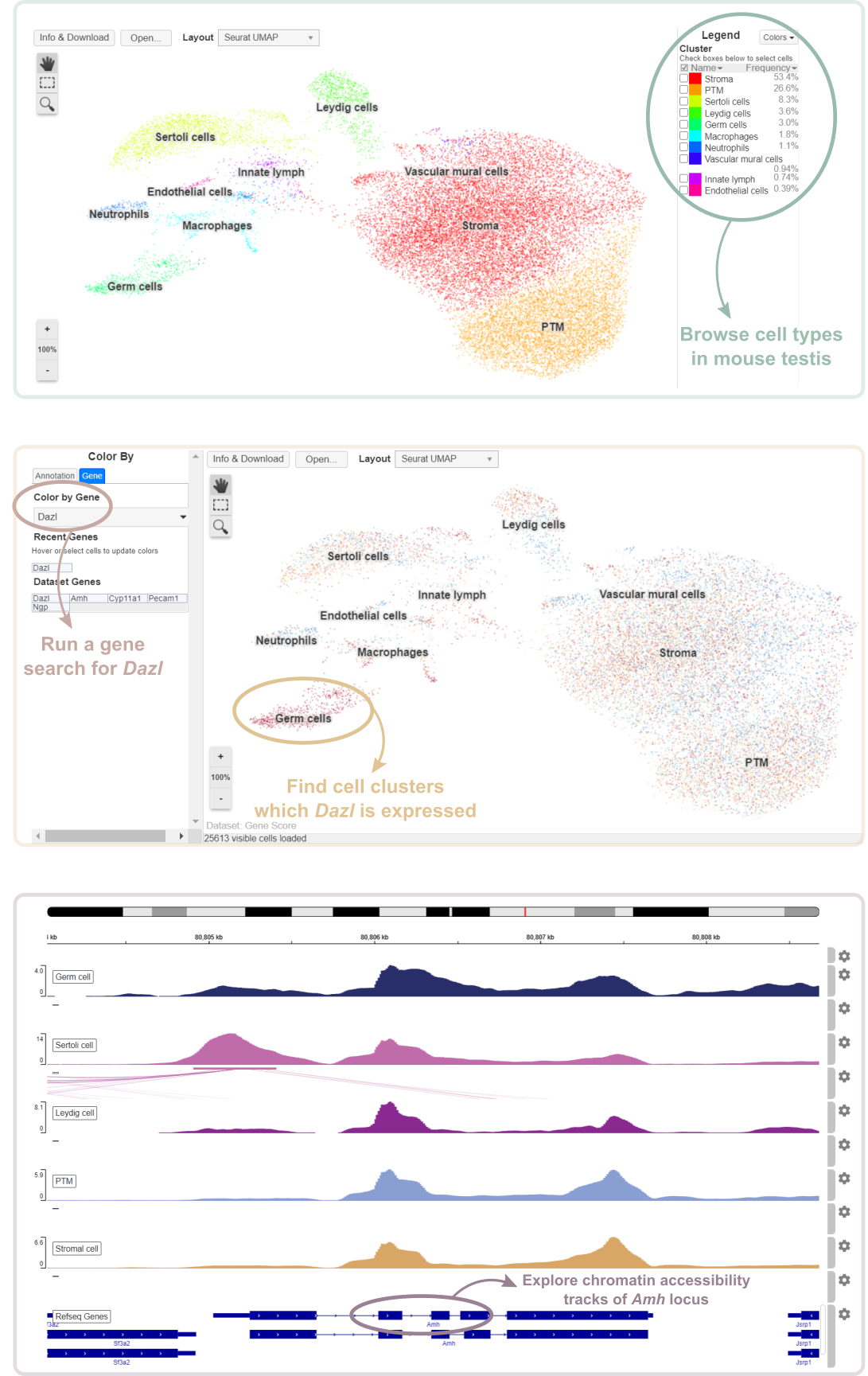
Usage examples
Citation
Jinyue Liao#, Hoi Ching Suen#, Shitao Rao, Alfred Chun Shui Luk, Ruoyu Zhang, Annie Wing Tung Lee, Ting Hei Thomas Chan, Man Yee Cheung, Ho Ting Chu, Hon Cheong So, Robin M. Hobbs, Tin Lap Lee. The single-cell epigenetic regulatory landscape in mammalian perinatal testis development. bioRxiv. doi: https://doi.org/10.1101/2021.03.17.435776